Docking to RNA via RMSD-driven Energy Minimization with Flexible Ligands and Flexible Targets
Christophe Guilbert and Thomas L. James
The following are Supplemental Materials to illustrate flexible docking using MORDOR.
Each image is actually a movie (gif animate)
which should automatically start when you open this web page , if
for some reasons, the image is static, you can directly click in
the image to download the movie in quick time format.
Docking Procedure
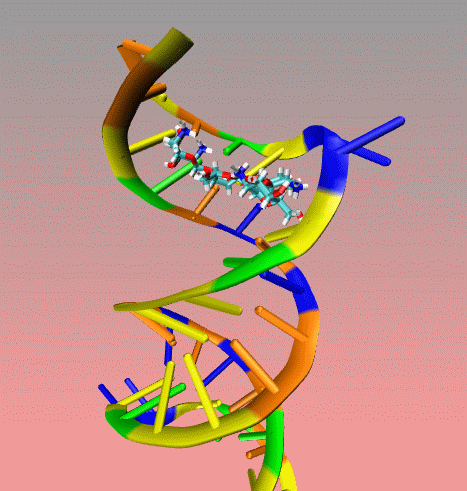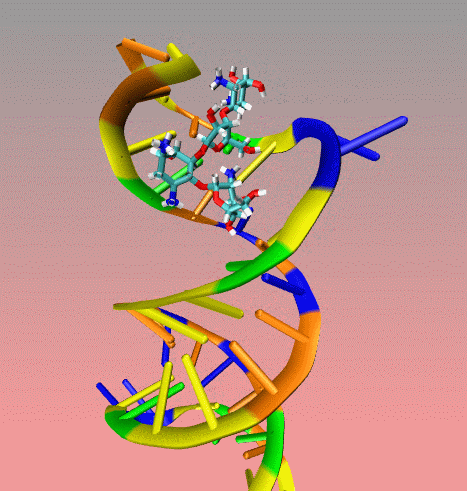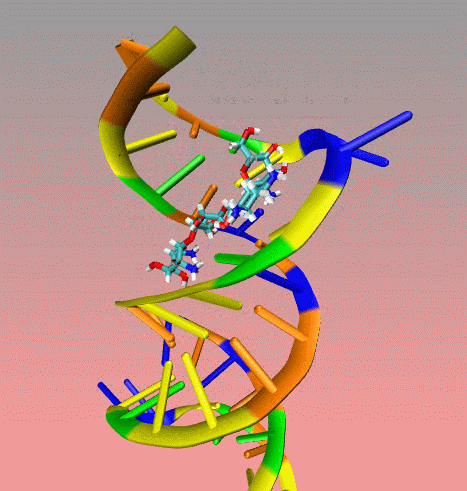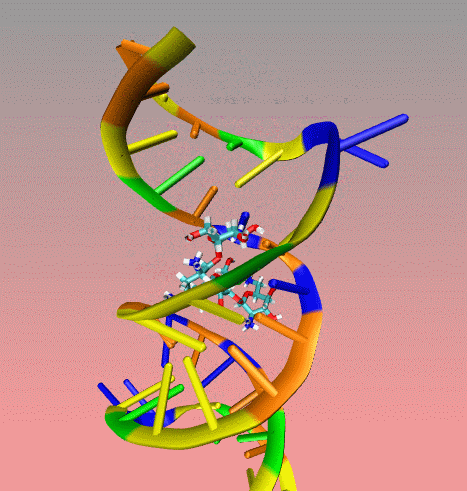 This
is an illustration of MORDOR docking procedure for the A-site on 16S
robosomal RNA from E. Coli complexed with the aminoglycoside
paromomycin (PDB code 1j7t). The following movie shows a few docking
steps "step 1 and 2" as descrived in the manuscript at the "
Docking via MORDOR" section.
This
is an illustration of MORDOR docking procedure for the A-site on 16S
robosomal RNA from E. Coli complexed with the aminoglycoside
paromomycin (PDB code 1j7t). The following movie shows a few docking
steps "step 1 and 2" as descrived in the manuscript at the "
Docking via MORDOR" section.
The first step consists of placing one
heavy atom of the ligand at one of the "hot spots" on the
receptor which are defined by a spheres (not seen). The orientation of
the ligand is defined by one of the 120 orientations uniformly
distributed on a trigonometric sphere. At this stage , the receptor is
held rigid. this stage can be easily identified in the movie since the
receptor is not moving. The 120 orientations are tested and
quickly minimized to remove any clash. Each time we find a "good"
orientation while trying all the possible orientations, we allow the
ligand to explore the receptor surface using PEDC (Path
Exploration with DIstance Constraints) algorithm (download article in pdf) enabling an induced fit of both the ligand and the receptor while the ligand probe the receptor.
This procedure is repeated 120 time for all the orientations and for all the heavy atoms in the ligand.
The following examples are not link to the manuscript but illustrate well the PEDC algotithm.
Example I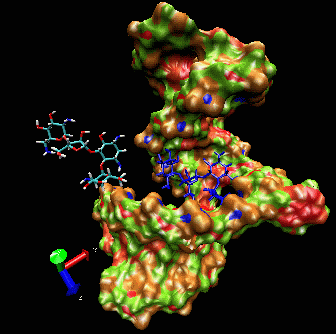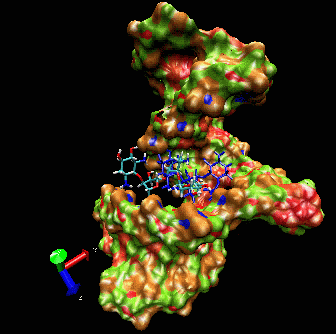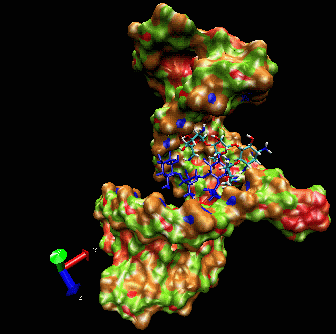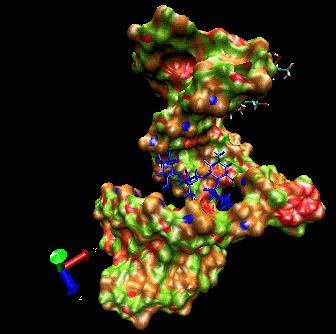
Here is a flexible
docking example using a region of the
ribosomal 16S RNA (pdb ID: 1fjf.pdb) to re-dock the antibiotic
paromomycin into its (known) binding pocket. The ligand in blue color
is the drug position as defined by the experimental X-ray structure.
Note the adaptation of the receptor as the ligand explores the groove.
The drug is “walking” along the surface mainly following
the major groove. One can clearly see that the ligand during the search
procedure binds specifically to the location found experimentally. The
ligand bound in this position has the best binding interaction energy.
However, it can still explore other possible binding sites.
Example II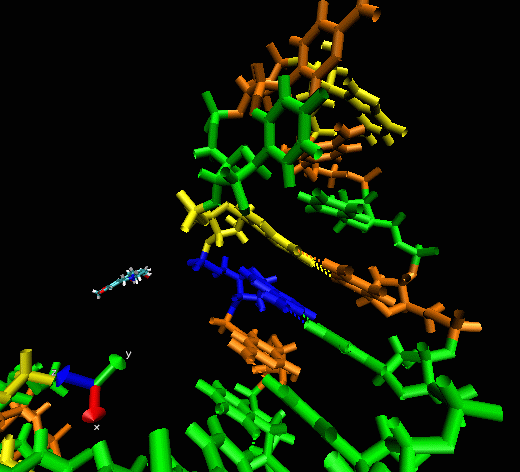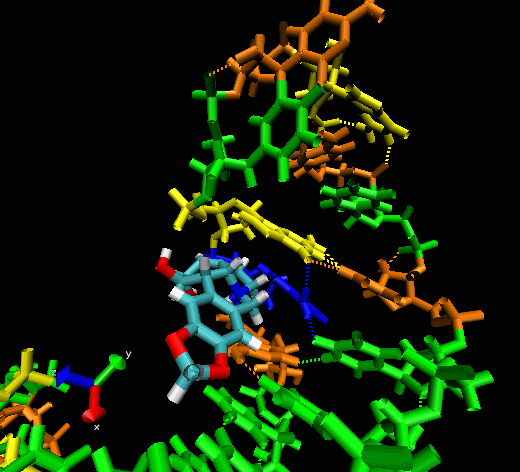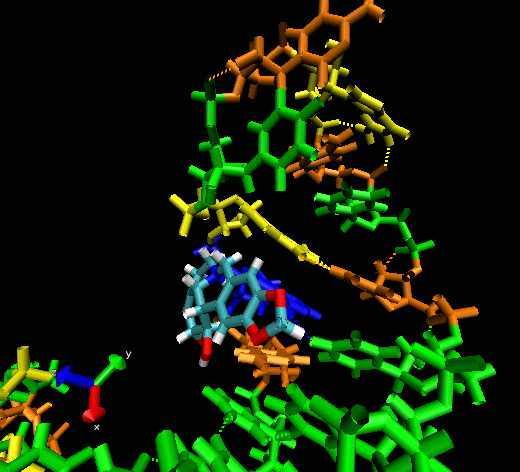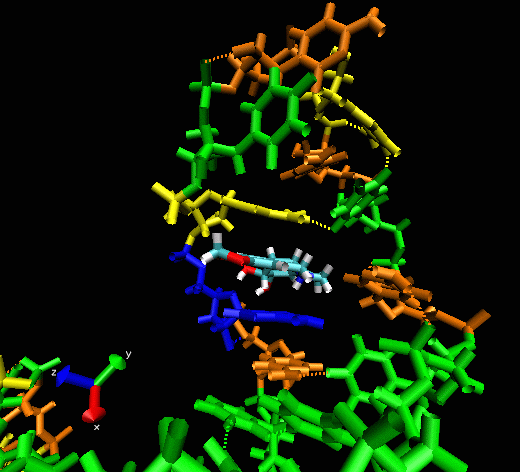
This example shows
the “induced fitting” ability of MORDOR when docking a
potential ligand to an RNA (telomerase) structure where there is no prior
experimental binding information. The ligand initially approaches one
of the sphere-identified “hot spots” (not seen) at the
surface of the RNA before exploring the macromolecular surface. One can
see that the compound briefly explores a pocket before (almost
instantaneously) intercalating between two bases, forming a nice
stacking interaction with them.
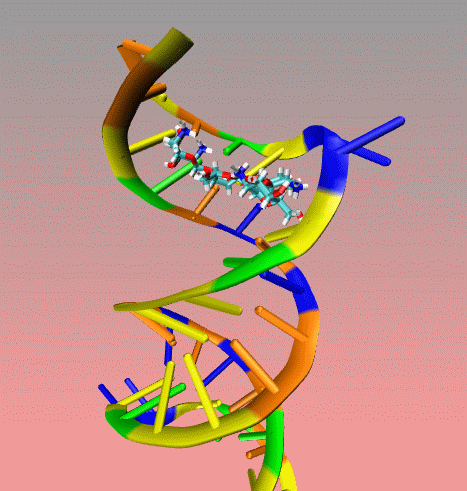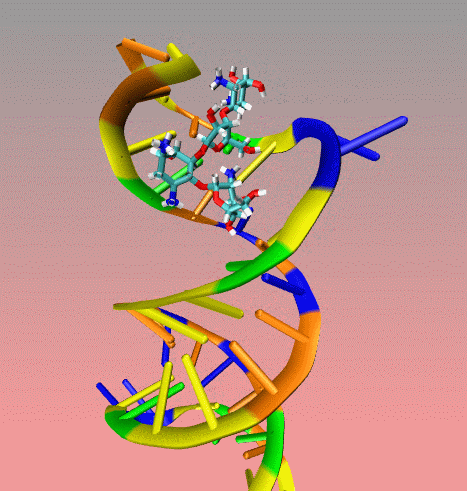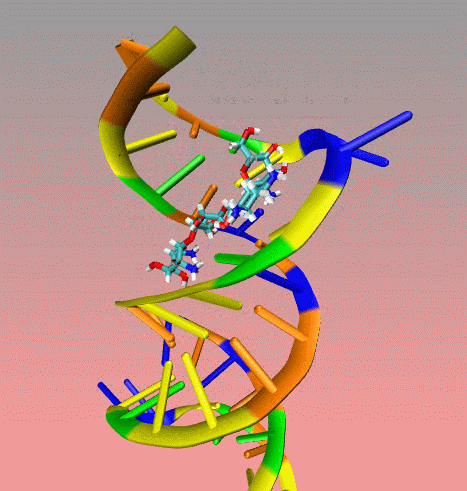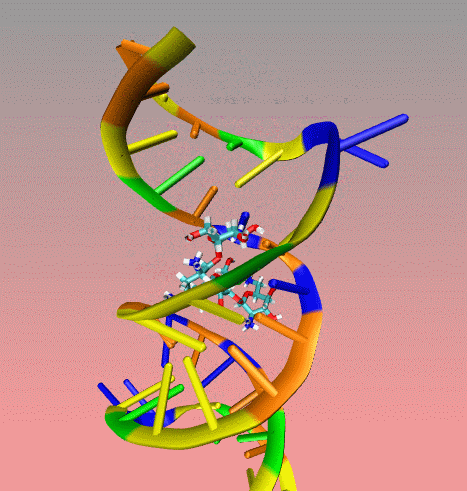 This
is an illustration of MORDOR docking procedure for the A-site on 16S
robosomal RNA from E. Coli complexed with the aminoglycoside
paromomycin (PDB code 1j7t). The following movie shows a few docking
steps "step 1 and 2" as descrived in the manuscript at the "
Docking via MORDOR" section.
This
is an illustration of MORDOR docking procedure for the A-site on 16S
robosomal RNA from E. Coli complexed with the aminoglycoside
paromomycin (PDB code 1j7t). The following movie shows a few docking
steps "step 1 and 2" as descrived in the manuscript at the "
Docking via MORDOR" section. 
